!lsREADME.md data docs index.qmd
_quarto.yml design.ipynb images seq_analysis.ipynbconda
docker
colab
To make google drive as colab working directory
from google.colab import drive
drive.mount('/content/drive')import os
os.chdir('/content/drive/MyDrive/design_build')!pwd
!wget https://media.addgene.org/snapgene-media/v1.7.9-0-g88a3305/sequences/222046/51c2cfab-a3b4-4d62-98df-0c77ec21164e/addgene-plasmid-50005-sequence-222046.gbk!lsREADME.md data docs index.qmd
_quarto.yml design.ipynb images seq_analysis.ipynb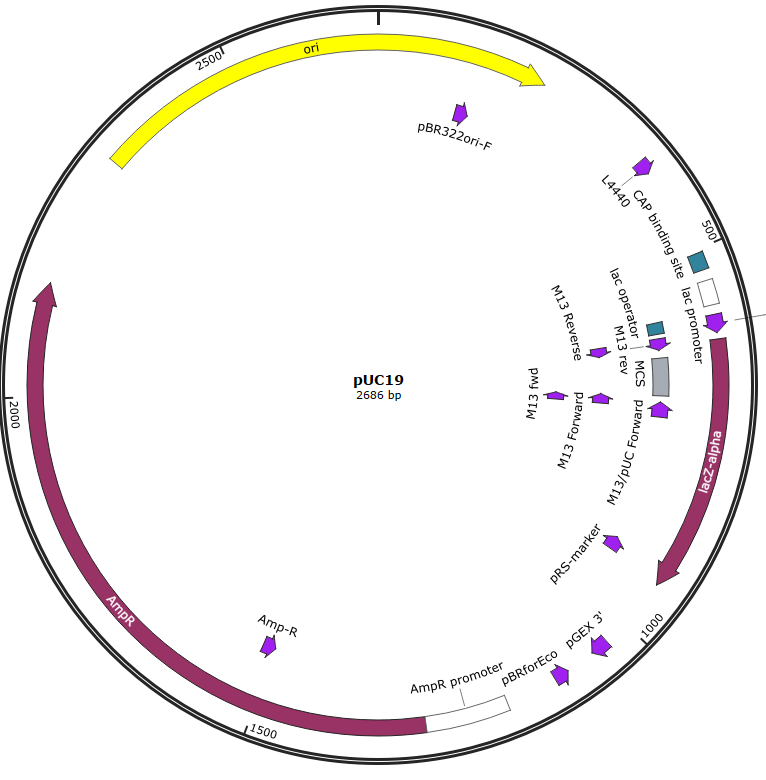
Biopython is a collection of freely available Python tools for computational biology and bioinformatics.
from Bio import SeqIO
from pandas import DataFrame
records = SeqIO.read("data/parts/addgene-plasmid-50005-sequence-222046.gbk", "genbank")
features = []
for feature in records.features:
features.append({
"Label": feature.qualifiers.get("label", [""])[0],
"Strand": feature.strand,
"Start": feature.location.start,
"End": feature.location.end,
"Type": feature.type
})
print(features)
DataFrame(features)[{'Label': '', 'Strand': 1, 'Start': ExactPosition(0), 'End': ExactPosition(2686), 'Type': 'source'}, {'Label': 'pBR322ori-F', 'Strand': 1, 'Start': ExactPosition(117), 'End': ExactPosition(137), 'Type': 'primer_bind'}, {'Label': 'L4440', 'Strand': 1, 'Start': ExactPosition(370), 'End': ExactPosition(388), 'Type': 'primer_bind'}, {'Label': 'CAP binding site', 'Strand': 1, 'Start': ExactPosition(504), 'End': ExactPosition(526), 'Type': 'protein_bind'}, {'Label': 'lac promoter', 'Strand': 1, 'Start': ExactPosition(540), 'End': ExactPosition(571), 'Type': 'promoter'}, {'Label': 'lac operator', 'Strand': 1, 'Start': ExactPosition(578), 'End': ExactPosition(595), 'Type': 'protein_bind'}, {'Label': 'M13/pUC Reverse', 'Strand': 1, 'Start': ExactPosition(583), 'End': ExactPosition(606), 'Type': 'primer_bind'}, {'Label': 'M13 rev', 'Strand': 1, 'Start': ExactPosition(602), 'End': ExactPosition(619), 'Type': 'primer_bind'}, {'Label': 'M13 Reverse', 'Strand': 1, 'Start': ExactPosition(602), 'End': ExactPosition(619), 'Type': 'primer_bind'}, {'Label': 'lacZ-alpha', 'Strand': 1, 'Start': ExactPosition(614), 'End': ExactPosition(938), 'Type': 'CDS'}, {'Label': 'MCS', 'Strand': 1, 'Start': ExactPosition(631), 'End': ExactPosition(688), 'Type': 'misc_feature'}, {'Label': 'M13 Forward', 'Strand': -1, 'Start': ExactPosition(688), 'End': ExactPosition(706), 'Type': 'primer_bind'}, {'Label': 'M13 fwd', 'Strand': -1, 'Start': ExactPosition(688), 'End': ExactPosition(705), 'Type': 'primer_bind'}, {'Label': 'M13/pUC Forward', 'Strand': -1, 'Start': ExactPosition(697), 'End': ExactPosition(720), 'Type': 'primer_bind'}, {'Label': 'pRS-marker', 'Strand': -1, 'Start': ExactPosition(913), 'End': ExactPosition(933), 'Type': 'primer_bind'}, {'Label': "pGEX 3'", 'Strand': 1, 'Start': ExactPosition(1032), 'End': ExactPosition(1055), 'Type': 'primer_bind'}, {'Label': 'pBRforEco', 'Strand': -1, 'Start': ExactPosition(1092), 'End': ExactPosition(1111), 'Type': 'primer_bind'}, {'Label': 'AmpR promoter', 'Strand': 1, 'Start': ExactPosition(1178), 'End': ExactPosition(1283), 'Type': 'promoter'}, {'Label': 'AmpR', 'Strand': 1, 'Start': ExactPosition(1283), 'End': ExactPosition(2144), 'Type': 'CDS'}, {'Label': 'Amp-R', 'Strand': -1, 'Start': ExactPosition(1501), 'End': ExactPosition(1521), 'Type': 'primer_bind'}, {'Label': 'ori', 'Strand': 1, 'Start': ExactPosition(0), 'End': ExactPosition(2686), 'Type': 'rep_origin'}]/home/haseong/anaconda3/envs/biopy/lib/python3.11/site-packages/Bio/SeqFeature.py:1040: BiopythonParserWarning: Attempting to fix invalid location '2315..217' as it looks like incorrect origin wrapping. Please fix input file, this could have unintended behavior.
warnings.warn(
/home/haseong/anaconda3/envs/biopy/lib/python3.11/site-packages/Bio/SeqFeature.py:230: BiopythonDeprecationWarning: Please use .location.strand rather than .strand
warnings.warn(| Label | Strand | Start | End | Type | |
|---|---|---|---|---|---|
| 0 | 1 | 0 | 2686 | source | |
| 1 | pBR322ori-F | 1 | 117 | 137 | primer_bind |
| 2 | L4440 | 1 | 370 | 388 | primer_bind |
| 3 | CAP binding site | 1 | 504 | 526 | protein_bind |
| 4 | lac promoter | 1 | 540 | 571 | promoter |
| 5 | lac operator | 1 | 578 | 595 | protein_bind |
| 6 | M13/pUC Reverse | 1 | 583 | 606 | primer_bind |
| 7 | M13 rev | 1 | 602 | 619 | primer_bind |
| 8 | M13 Reverse | 1 | 602 | 619 | primer_bind |
| 9 | lacZ-alpha | 1 | 614 | 938 | CDS |
| 10 | MCS | 1 | 631 | 688 | misc_feature |
| 11 | M13 Forward | -1 | 688 | 706 | primer_bind |
| 12 | M13 fwd | -1 | 688 | 705 | primer_bind |
| 13 | M13/pUC Forward | -1 | 697 | 720 | primer_bind |
| 14 | pRS-marker | -1 | 913 | 933 | primer_bind |
| 15 | pGEX 3' | 1 | 1032 | 1055 | primer_bind |
| 16 | pBRforEco | -1 | 1092 | 1111 | primer_bind |
| 17 | AmpR promoter | 1 | 1178 | 1283 | promoter |
| 18 | AmpR | 1 | 1283 | 2144 | CDS |
| 19 | Amp-R | -1 | 1501 | 1521 | primer_bind |
| 20 | ori | 1 | 0 | 2686 | rep_origin |
primers It is uniquely focused on DNA assembly flows like Gibson Assembly and Golden Gate cloning. You can design primers while adding sequence to the 5’ ends of primers.
from primers import create, primers
from pandas import DataFrame
from random import sample, choices
myseq_list = choices(["A", "T", "G", "C"], k=100)
myseq = "".join(myseq_list)
print(myseq)
fwd, rev = create(myseq, add_fwd = "GGGG", add_rev = "TTTT")
# p1, p2 = primers(myseq, add_fwd = "GGGG", add_rev = "TTTT")
print(fwd)
print(rev)
## display table form
DataFrame(list(fwd.dict().values())[:-1], index = list(rev.dict().keys())[:-1])
## default argument values
createCGTGCCTCCATAAATAACTTGCAAGATTCTCACCATTCGAAGGTTCTCGACAAGGGGCGGGGGGTAAAAATAGCATTACTAGTTCGGATAAATCTGCCCT
Primer(seq='GGGGCGTGCCTCCATAAATAACTTG', len=25, tm=63.3, tm_total=70.6, gc=0.5, dg=0, fwd=True, off_target_count=0, scoring=Scoring(penalty=1.8, penalty_tm=1.3, penalty_tm_diff=0, penalty_gc=0.0, penalty_len=0.5, penalty_dg=0.0, penalty_off_target=0.0))
Primer(seq='TTTTAGGGCAGATTTATCCGAACTAGT', len=27, tm=63.4, tm_total=63.9, gc=0.4, dg=-0.37, fwd=False, off_target_count=0, scoring=Scoring(penalty=4.6, penalty_tm=1.4, penalty_tm_diff=0, penalty_gc=2.0, penalty_len=0.5, penalty_dg=0.7, penalty_off_target=0.0))<function primers.primers.primers(seq: str, add_fwd: str = '', add_rev: str = '', add_fwd_len: Tuple[int, int] = (-1, -1), add_rev_len: Tuple[int, int] = (-1, -1), offtarget_check: str = '', optimal_tm: float = 62.0, optimal_gc: float = 0.5, optimal_len: int = 22, penalty_tm: float = 1.0, penalty_gc: float = 0.2, penalty_len: float = 0.5, penalty_tm_diff: float = 1.0, penalty_dg: float = 2.0, penalty_off_target: float = 20.0) -> Tuple[primers.primers.Primer, primers.primers.Primer]><function primers.primers.primers(seq: str, add_fwd: str = '', add_rev: str = '', add_fwd_len: Tuple[int, int] = (-1, -1), add_rev_len: Tuple[int, int] = (-1, -1), offtarget_check: str = '', optimal_tm: float = 62.0, optimal_gc: float = 0.5, optimal_len: int = 22, penalty_tm: float = 1.0, penalty_gc: float = 0.2, penalty_len: float = 0.5, penalty_tm_diff: float = 1.0, penalty_dg: float = 2.0, penalty_off_target: float = 20.0) -> Tuple[primers.primers.Primer, primers.primers.Primer]>from primers import create
from random import choices
def print_primer_info(x):
from pandas import DataFrame
df = DataFrame(list(x.dict().values())[:-1], index = list(x.dict().keys())[:-1])
print(df)
primer_binding_seq = "GTCATATGCATTCGATGCGTTAGG"
rnd_seq1 = "".join(choices(["A", "T", "G", "C"], k=100))
rnd_seq2 = "".join(choices(["A", "T", "G", "C"], k=100))
myseq = primer_binding_seq+rnd_seq1
print(myseq)
len(myseq)
fwd, rev = create(myseq)
print_primer_info(fwd)
## primer considering offtargets
myseq2 = primer_binding_seq+rnd_seq1+primer_binding_seq+rnd_seq2
fwd2, rev = create(myseq2)
print_primer_info(fwd2)
## optimal_tm is ignored
fwd2, rev = create(myseq2, optimal_tm = 62)
print_primer_info(fwd2)GTCATATGCATTCGATGCGTTAGGGACACCCATGGCAACATGTGGATATAACTCGGTGCTGAGGAAAACTTCATACGCTCTTGACGTTCTATGCATGAAGCCCTTTCAACGCACGCTTCATTCA
0
seq GTCATATGCATTCGATGCGT
len 20
tm 62.5
tm_total 62.5
gc 0.5
dg -0.56
fwd True
off_target_count 0
0
seq GTCATATGCATTCGATGCGTTAGGGA
len 26
tm 68.0
tm_total 68.0
gc 0.5
dg -0.56
fwd True
off_target_count 0
0
seq GTCATATGCATTCGATGCGTTAGGGA
len 26
tm 68.0
tm_total 68.0
gc 0.5
dg -0.56
fwd True
off_target_count 0from pydna.dseqrecord import Dseqrecord
dsr = Dseqrecord("ATGCGTTGC")
dsr.figure()Dseqrecord(-9) ATGCGTTGC TACGCAACG
from pydna.readers import read
p = read("data/parts/addgene-plasmid-50005-sequence-222046.gbk")
p.list_features()/home/haseong/anaconda3/envs/biopy/lib/python3.11/site-packages/Bio/SeqFeature.py:1040: BiopythonParserWarning: Attempting to fix invalid location '2315..217' as it looks like incorrect origin wrapping. Please fix input file, this could have unintended behavior.
warnings.warn(| Ft# | Label or Note | Dir | Sta | End | Len | type | orf? |
|---|---|---|---|---|---|---|---|
| 0 | nd | --> | 0 | 2686 | 2686 | source | no |
| 1 | L:pBR322ori-F | --> | 117 | 137 | 20 | primer_bind | no |
| 2 | L:L4440 | --> | 370 | 388 | 18 | primer_bind | no |
| 3 | L:CAP binding si | --> | 504 | 526 | 22 | protein_bind | no |
| 4 | L:lac promoter | --> | 540 | 571 | 31 | promoter | no |
| 5 | L:lac operator | --> | 578 | 595 | 17 | protein_bind | no |
| 6 | L:M13/pUC Revers | --> | 583 | 606 | 23 | primer_bind | no |
| 7 | L:M13 rev | --> | 602 | 619 | 17 | primer_bind | no |
| 8 | L:M13 Reverse | --> | 602 | 619 | 17 | primer_bind | no |
| 9 | L:lacZ-alpha | --> | 614 | 938 | 324 | CDS | yes |
| 10 | L:MCS | --> | 631 | 688 | 57 | misc_feature | no |
| 11 | L:M13 Forward | <-- | 688 | 706 | 18 | primer_bind | no |
| 12 | L:M13 fwd | <-- | 688 | 705 | 17 | primer_bind | no |
| 13 | L:M13/pUC Forwar | <-- | 697 | 720 | 23 | primer_bind | no |
| 14 | L:pRS-marker | <-- | 913 | 933 | 20 | primer_bind | no |
| 15 | L:pGEX 3' | --> | 1032 | 1055 | 23 | primer_bind | no |
| 16 | L:pBRforEco | <-- | 1092 | 1111 | 19 | primer_bind | no |
| 17 | L:AmpR promoter | --> | 1178 | 1283 | 105 | promoter | no |
| 18 | L:AmpR | --> | 1283 | 2144 | 861 | CDS | yes |
| 19 | L:Amp-R | <-- | 1501 | 1521 | 20 | primer_bind | no |
| 20 | L:ori | --> | 0 | 2686 | 589 | rep_origin | no |
extracted_site = p.extract_feature(10)
extracted_site.seqDseq(-57)
AAGC..ATTC
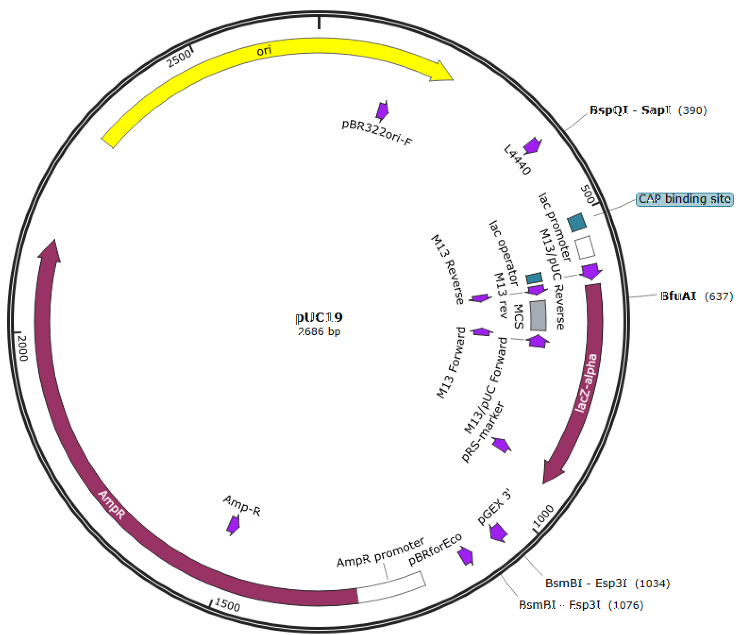
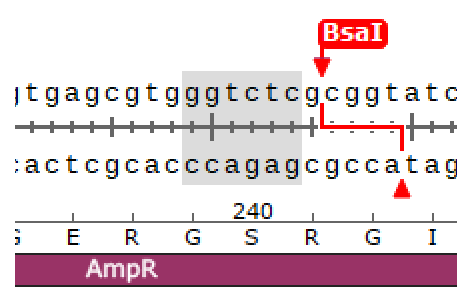
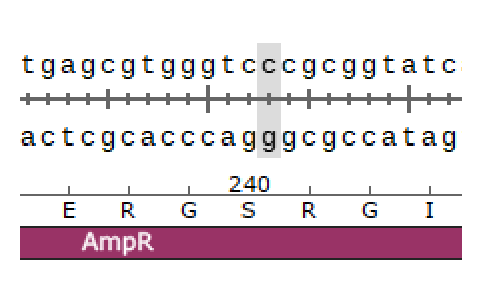
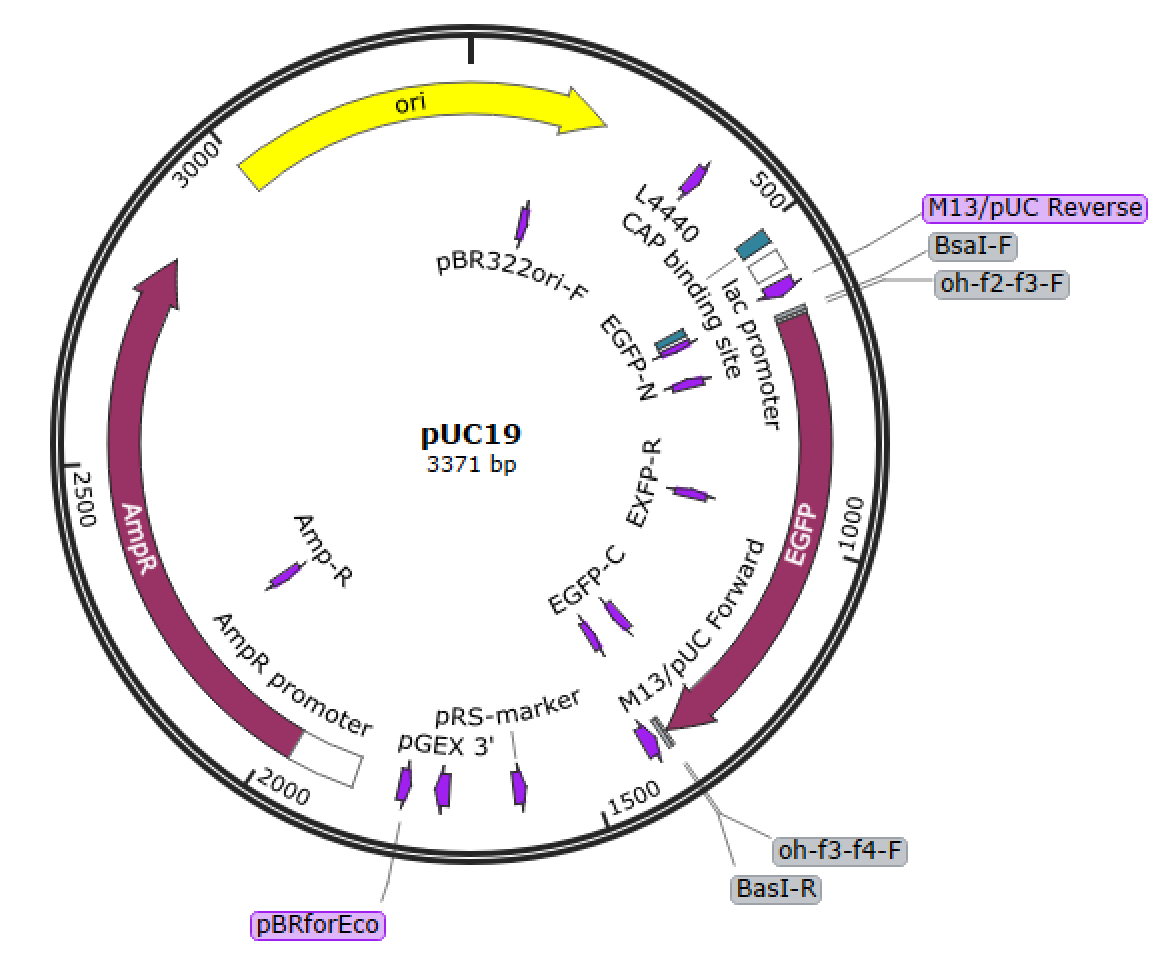
TTCG..TAAGpUC19 from Addgene
Remove BsaI site
Insert a part into MCS
Hinz, Aaron J., Benjamin Stenzler, and Alexandre J. Poulain. “Golden gate assembly of aerobic and anaerobic microbial bioreporters.” Applied and environmental microbiology 88.1 (2022): e01485-21.
from pydna.readers import read
from primers import create
egfp = read("data/parts/pUC19-egfp.gb")
egfp_feature_list = egfp.list_features()
display(egfp_feature_list)
myseq = egfp.extract_feature(13)
str(myseq.seq)
fwd, rev = create(str(myseq.seq), add_fwd = "GGGG", add_rev = "TTTT")
print(fwd)/home/haseong/anaconda3/envs/biopy/lib/python3.11/site-packages/Bio/GenBank/Scanner.py:1528: BiopythonParserWarning: Attempting to parse malformed locus line:
'LOCUS pUC19-egfp 3371 bp DNA circular SYN 18-MAY-2024\n'
Found locus 'pUC19-egfp' size '3371' residue_type 'DNA'
Some fields may be wrong.
warnings.warn(| Ft# | Label or Note | Dir | Sta | End | Len | type | orf? |
|---|---|---|---|---|---|---|---|
| 0 | nd | --> | 0 | 3371 | 3371 | source | no |
| 1 | L:pBR322ori-F | --> | 117 | 137 | 20 | primer_bind | no |
| 2 | L:L4440 | --> | 370 | 388 | 18 | primer_bind | no |
| 3 | L:CAP binding si | --> | 504 | 526 | 22 | protein_bind | no |
| 4 | L:lac promoter | --> | 540 | 571 | 31 | promoter | no |
| 5 | L:lac operator | --> | 578 | 595 | 17 | protein_bind | no |
| 6 | L:M13/pUC Revers | --> | 583 | 606 | 23 | primer_bind | no |
| 7 | L:M13 rev | --> | 602 | 619 | 17 | primer_bind | no |
| 8 | L:M13 Reverse | --> | 602 | 619 | 17 | primer_bind | no |
| 9 | L:lacZ-alpha | --> | 614 | 1623 | 1009 | CDS | no |
| 10 | L:BsaI-F | --> | 631 | 638 | 7 | misc_feature | no |
| 11 | L:lacZ-alpha | --> | 638 | 642 | 4 | CDS | no |
| 12 | L:oh-f2-f3-F | --> | 638 | 642 | 4 | misc_feature | no |
| 13 | L:EGFP | --> | 642 | 1362 | 720 | CDS | yes |
| 14 | L:EGFP | --> | 642 | 1359 | 717 | CDS | no |
| 15 | L:EGFP-N | <-- | 687 | 709 | 22 | primer_bind | no |
| 16 | L:EXFP-R | <-- | 948 | 968 | 20 | primer_bind | no |
| 17 | L:EGFP-C | --> | 1295 | 1317 | 22 | primer_bind | no |
| 18 | L:oh-f3-f4-F | --> | 1362 | 1366 | 4 | misc_feature | no |
| 19 | L:BasI-R | --> | 1366 | 1373 | 7 | misc_feature | no |
| 20 | L:M13 Forward | <-- | 1373 | 1391 | 18 | primer_bind | no |
| 21 | L:M13 fwd | <-- | 1373 | 1390 | 17 | primer_bind | no |
| 22 | L:M13/pUC Forwar | <-- | 1382 | 1405 | 23 | primer_bind | no |
| 23 | L:pRS-marker | <-- | 1598 | 1618 | 20 | primer_bind | no |
| 24 | L:pGEX 3' | --> | 1717 | 1740 | 23 | primer_bind | no |
| 25 | L:pBRforEco | <-- | 1777 | 1796 | 19 | primer_bind | no |
| 26 | L:AmpR promoter | --> | 1863 | 1968 | 105 | promoter | no |
| 27 | L:AmpR | --> | 1968 | 2829 | 861 | CDS | yes |
| 28 | L:Amp-R | <-- | 2186 | 2206 | 20 | primer_bind | no |
| 29 | L:ori | --> | 0 | 3371 | 589 | rep_origin | no |
Primer(seq='GGGGATGGTGAGCAAGGGCG', len=20, tm=65.1, tm_total=72.1, gc=0.7, dg=0, fwd=True, off_target_count=0, scoring=Scoring(penalty=10.1, penalty_tm=3.1, penalty_tm_diff=0, penalty_gc=4.0, penalty_len=3.0, penalty_dg=0.0, penalty_off_target=0.0))from pydna.all import pcr
pcr_product = pcr(fwd.seq, rev.seq, egfp.extract_feature(13))
pcr_product.figure() 5ATGGTGAGCAAGGGCG...CATGGACGAGCTGTACAAGTAA3
||||||||||||||||||||||
3GTACCTGCTCGACATGTTCATTTTTT5
5GGGGATGGTGAGCAAGGGCG3
||||||||||||||||
3TACCACTCGTTCCCGC...GTACCTGCTCGACATGTTCATT5from primers import create
from pydna.all import pcr
frag1 = egfp.extract_feature(10)
frag2 = egfp.extract_feature(12)
frag3 = egfp.extract_feature(13)
frag4 = egfp.extract_feature(18)
frag5 = egfp.extract_feature(19)
targetseq = frag1+frag2+frag3+frag4+frag5
fwd, rev = create(str(targetseq.seq))
pcr_product = pcr(fwd.seq, rev.seq, targetseq)
pcr_product.figure()5GGTCTCAGTCAATGGTGA...TACAAGTAAGGGATGAGACC3
||||||||||||||||||||
3ATGTTCATTCCCTACTCTGG5
5GGTCTCAGTCAATGGTGA3
||||||||||||||||||
3CCAGAGTCAGTTACCACT...ATGTTCATTCCCTACTCTGG5from Bio.Restriction import BsaI
cut_product = pcr_product.cut(BsaI)
print(len(cut_product))
display(cut_product[0].figure())
print()
display(cut_product[1].figure())
print()
display(cut_product[2].figure())3
Dseqrecord(-11)
GGTCTCA
CCAGAGTCAGT
Dseqrecord(-728)
GTCAATGGTGAGCAAGGGCGAGGAGCTGTTCACCGGGGTGGTGCCCATCCTGGTCGAGCTGGACGGCGACGTAAACGGCCACAAGTTCAGCGTGTCCGGCGAGGGCGAGGGCGATGCCACCTACGGCAAGCTGACCCTGAAGTTCATCTGCACCACCGGCAAGCTGCCCGTGCCCTGGCCCACCCTCGTGACCACCCTGACCTACGGCGTGCAGTGCTTCAGCCGCTACCCCGACCACATGAAGCAGCACGACTTCTTCAAGTCCGCCATGCCCGAAGGCTACGTCCAGGAGCGCACCATCTTCTTCAAGGACGACGGCAACTACAAGACCCGCGCCGAGGTGAAGTTCGAGGGCGACACCCTGGTGAACCGCATCGAGCTGAAGGGCATCGACTTCAAGGAGGACGGCAACATCCTGGGGCACAAGCTGGAGTACAACTACAACAGCCACAACGTCTATATCATGGCCGACAAGCAGAAGAACGGCATCAAGGTGAACTTCAAGATCCGCCACAACATCGAGGACGGCAGCGTGCAGCTCGCCGACCACTACCAGCAGAACACCCCCATCGGCGACGGCCCCGTGCTGCTGCCCGACAACCACTACCTGAGCACCCAGTCCGCCCTGAGCAAAGACCCCAACGAGAAGCGCGATCACATGGTCCTGCTGGAGTTCGTGACCGCCGCCGGGATCACTCTCGGCATGGACGAGCTGTACAAGTAA
TACCACTCGTTCCCGCTCCTCGACAAGTGGCCCCACCACGGGTAGGACCAGCTCGACCTGCCGCTGCATTTGCCGGTGTTCAAGTCGCACAGGCCGCTCCCGCTCCCGCTACGGTGGATGCCGTTCGACTGGGACTTCAAGTAGACGTGGTGGCCGTTCGACGGGCACGGGACCGGGTGGGAGCACTGGTGGGACTGGATGCCGCACGTCACGAAGTCGGCGATGGGGCTGGTGTACTTCGTCGTGCTGAAGAAGTTCAGGCGGTACGGGCTTCCGATGCAGGTCCTCGCGTGGTAGAAGAAGTTCCTGCTGCCGTTGATGTTCTGGGCGCGGCTCCACTTCAAGCTCCCGCTGTGGGACCACTTGGCGTAGCTCGACTTCCCGTAGCTGAAGTTCCTCCTGCCGTTGTAGGACCCCGTGTTCGACCTCATGTTGATGTTGTCGGTGTTGCAGATATAGTACCGGCTGTTCGTCTTCTTGCCGTAGTTCCACTTGAAGTTCTAGGCGGTGTTGTAGCTCCTGCCGTCGCACGTCGAGCGGCTGGTGATGGTCGTCTTGTGGGGGTAGCCGCTGCCGGGGCACGACGACGGGCTGTTGGTGATGGACTCGTGGGTCAGGCGGGACTCGTTTCTGGGGTTGCTCTTCGCGCTAGTGTACCAGGACGACCTCAAGCACTGGCGGCGGCCCTAGTGAGAGCCGTACCTGCTCGACATGTTCATTCCCT
Dseqrecord(-11)
GGGATGAGACC
ACTCTGG
from pydna.assembly import Assembly
from pydna.common_sub_strings import terminal_overlap
asm = Assembly((cut_product[0], cut_product[1], cut_product[2]), algorithm = terminal_overlap, limit = 4)
candidate = asm.assemble_linear()
print(candidate[0])
candidate[0].figure() Dseqrecord
circular: False
size: 742
ID: id
Name: name
Description: description
Number of features: 13
/molecule_type=DNA
Dseq(-742)
GGTC..GACC
CCAG..CTGGname| 4
\/
/\
4|name| 4
\/
/\
4|namefrom pydna.readers import read
## read the parts
egfp = read("data/parts/pUC19-egfp.gb")
promoter = read("data/parts/pUC19-J23100.gb")
terminator = read("data/parts/pUC19-L2U3H03.gb")
rbs = read("data/parts/pUC19-RB0030.gb")
display(promoter.list_features())
display(rbs.list_features())
display(egfp.list_features())
display(terminator.list_features())/home/haseong/anaconda3/envs/biopy/lib/python3.11/site-packages/Bio/GenBank/Scanner.py:1528: BiopythonParserWarning: Attempting to parse malformed locus line:
'LOCUS pUC19-egfp 3371 bp DNA circular SYN 18-MAY-2024\n'
Found locus 'pUC19-egfp' size '3371' residue_type 'DNA'
Some fields may be wrong.
warnings.warn(
/home/haseong/anaconda3/envs/biopy/lib/python3.11/site-packages/Bio/GenBank/Scanner.py:1528: BiopythonParserWarning: Attempting to parse malformed locus line:
'LOCUS pUC19-J23100 2686 bp DNA circular SYN 01-JUN-2024\n'
Found locus 'pUC19-J23100' size '2686' residue_type 'DNA'
Some fields may be wrong.
warnings.warn(
/home/haseong/anaconda3/envs/biopy/lib/python3.11/site-packages/Bio/GenBank/Scanner.py:1528: BiopythonParserWarning: Attempting to parse malformed locus line:
'LOCUS pUC19-L2U3H03 2688 bp DNA circular SYN 01-JUN-2024\n'
Found locus 'pUC19-L2U3H03' size '2688' residue_type 'DNA'
Some fields may be wrong.
warnings.warn(
/home/haseong/anaconda3/envs/biopy/lib/python3.11/site-packages/Bio/GenBank/Scanner.py:1528: BiopythonParserWarning: Attempting to parse malformed locus line:
'LOCUS pUC19-RB0030 2750 bp DNA circular SYN 01-JUN-2024\n'
Found locus 'pUC19-RB0030' size '2750' residue_type 'DNA'
Some fields may be wrong.
warnings.warn(| Ft# | Label or Note | Dir | Sta | End | Len | type | orf? |
|---|---|---|---|---|---|---|---|
| 0 | nd | --> | 0 | 2686 | 2686 | source | no |
| 1 | L:pBR322ori-F | --> | 117 | 137 | 20 | primer_bind | no |
| 2 | L:L4440 | --> | 370 | 388 | 18 | primer_bind | no |
| 3 | L:CAP binding si | --> | 504 | 526 | 22 | protein_bind | no |
| 4 | L:lac promoter | --> | 540 | 571 | 31 | promoter | no |
| 5 | L:lac operator | --> | 578 | 595 | 17 | protein_bind | no |
| 6 | L:M13/pUC Revers | --> | 583 | 606 | 23 | primer_bind | no |
| 7 | L:M13 rev | --> | 602 | 619 | 17 | primer_bind | no |
| 8 | L:M13 Reverse | --> | 602 | 619 | 17 | primer_bind | no |
| 9 | L:lacZ-alpha | --> | 614 | 938 | 324 | CDS | no |
| 10 | L:BsaI-F | --> | 631 | 638 | 7 | misc_feature | no |
| 11 | L:oh-v-f1-F | --> | 638 | 642 | 4 | misc_feature | no |
| 12 | L:BBa_J23100 | --> | 642 | 677 | 35 | promoter | no |
| 13 | L:oh-f1-f2-F | --> | 677 | 681 | 4 | misc_feature | no |
| 14 | L:BsaI-R | --> | 681 | 688 | 7 | misc_feature | no |
| 15 | L:M13 Forward | <-- | 688 | 706 | 18 | primer_bind | no |
| 16 | L:M13 fwd | <-- | 688 | 705 | 17 | primer_bind | no |
| 17 | L:M13/pUC Forwar | <-- | 697 | 720 | 23 | primer_bind | no |
| 18 | L:pRS-marker | <-- | 913 | 933 | 20 | primer_bind | no |
| 19 | L:pGEX 3' | --> | 1032 | 1055 | 23 | primer_bind | no |
| 20 | L:pBRforEco | <-- | 1092 | 1111 | 19 | primer_bind | no |
| 21 | L:AmpR promoter | --> | 1178 | 1283 | 105 | promoter | no |
| 22 | L:AmpR | --> | 1283 | 2144 | 861 | CDS | yes |
| 23 | L:Amp-R | <-- | 1501 | 1521 | 20 | primer_bind | no |
| 24 | L:ori | --> | 0 | 2686 | 589 | rep_origin | no |
| Ft# | Label or Note | Dir | Sta | End | Len | type | orf? |
|---|---|---|---|---|---|---|---|
| 0 | nd | --> | 0 | 2750 | 2750 | source | no |
| 1 | L:pBR322ori-F | --> | 117 | 137 | 20 | primer_bind | no |
| 2 | L:L4440 | --> | 370 | 388 | 18 | primer_bind | no |
| 3 | L:CAP binding si | --> | 504 | 526 | 22 | protein_bind | no |
| 4 | L:lac promoter | --> | 540 | 571 | 31 | promoter | no |
| 5 | L:lac operator | --> | 578 | 595 | 17 | protein_bind | no |
| 6 | L:M13/pUC Revers | --> | 583 | 606 | 23 | primer_bind | no |
| 7 | L:M13 rev | --> | 602 | 619 | 17 | primer_bind | no |
| 8 | L:M13 Reverse | --> | 602 | 619 | 17 | primer_bind | no |
| 9 | L:BsaI-F | --> | 631 | 638 | 7 | misc_feature | no |
| 10 | L:oh-f1-f2-F | --> | 638 | 642 | 4 | misc_feature | no |
| 11 | L:R_BBa_B0030 | --> | 642 | 741 | 99 | misc_feature | no |
| 12 | L:sTRSV HHRz | --> | 644 | 694 | 50 | misc_RNA | no |
| 13 | N:strong bacteri | --> | 727 | 739 | 12 | RBS | no |
| 14 | L:oh-f2-f3-F | --> | 741 | 745 | 4 | misc_feature | no |
| 15 | L:BsaI-R | --> | 745 | 752 | 7 | misc_feature | no |
| 16 | L:M13 Forward | <-- | 752 | 770 | 18 | primer_bind | no |
| 17 | L:M13 fwd | <-- | 752 | 769 | 17 | primer_bind | no |
| 18 | L:M13/pUC Forwar | <-- | 761 | 784 | 23 | primer_bind | no |
| 19 | L:pRS-marker | <-- | 977 | 997 | 20 | primer_bind | no |
| 20 | L:pGEX 3' | --> | 1096 | 1119 | 23 | primer_bind | no |
| 21 | L:pBRforEco | <-- | 1156 | 1175 | 19 | primer_bind | no |
| 22 | L:AmpR promoter | --> | 1242 | 1347 | 105 | promoter | no |
| 23 | L:AmpR | --> | 1347 | 2208 | 861 | CDS | yes |
| 24 | L:Amp-R | <-- | 1565 | 1585 | 20 | primer_bind | no |
| 25 | L:ori | --> | 0 | 2750 | 589 | rep_origin | no |
| Ft# | Label or Note | Dir | Sta | End | Len | type | orf? |
|---|---|---|---|---|---|---|---|
| 0 | nd | --> | 0 | 3371 | 3371 | source | no |
| 1 | L:pBR322ori-F | --> | 117 | 137 | 20 | primer_bind | no |
| 2 | L:L4440 | --> | 370 | 388 | 18 | primer_bind | no |
| 3 | L:CAP binding si | --> | 504 | 526 | 22 | protein_bind | no |
| 4 | L:lac promoter | --> | 540 | 571 | 31 | promoter | no |
| 5 | L:lac operator | --> | 578 | 595 | 17 | protein_bind | no |
| 6 | L:M13/pUC Revers | --> | 583 | 606 | 23 | primer_bind | no |
| 7 | L:M13 rev | --> | 602 | 619 | 17 | primer_bind | no |
| 8 | L:M13 Reverse | --> | 602 | 619 | 17 | primer_bind | no |
| 9 | L:lacZ-alpha | --> | 614 | 1623 | 1009 | CDS | no |
| 10 | L:BsaI-F | --> | 631 | 638 | 7 | misc_feature | no |
| 11 | L:lacZ-alpha | --> | 638 | 642 | 4 | CDS | no |
| 12 | L:oh-f2-f3-F | --> | 638 | 642 | 4 | misc_feature | no |
| 13 | L:EGFP | --> | 642 | 1362 | 720 | CDS | yes |
| 14 | L:EGFP | --> | 642 | 1359 | 717 | CDS | no |
| 15 | L:EGFP-N | <-- | 687 | 709 | 22 | primer_bind | no |
| 16 | L:EXFP-R | <-- | 948 | 968 | 20 | primer_bind | no |
| 17 | L:EGFP-C | --> | 1295 | 1317 | 22 | primer_bind | no |
| 18 | L:oh-f3-f4-F | --> | 1362 | 1366 | 4 | misc_feature | no |
| 19 | L:BsaI-R | --> | 1366 | 1373 | 7 | misc_feature | no |
| 20 | L:M13 Forward | <-- | 1373 | 1391 | 18 | primer_bind | no |
| 21 | L:M13 fwd | <-- | 1373 | 1390 | 17 | primer_bind | no |
| 22 | L:M13/pUC Forwar | <-- | 1382 | 1405 | 23 | primer_bind | no |
| 23 | L:pRS-marker | <-- | 1598 | 1618 | 20 | primer_bind | no |
| 24 | L:pGEX 3' | --> | 1717 | 1740 | 23 | primer_bind | no |
| 25 | L:pBRforEco | <-- | 1777 | 1796 | 19 | primer_bind | no |
| 26 | L:AmpR promoter | --> | 1863 | 1968 | 105 | promoter | no |
| 27 | L:AmpR | --> | 1968 | 2829 | 861 | CDS | yes |
| 28 | L:Amp-R | <-- | 2186 | 2206 | 20 | primer_bind | no |
| 29 | L:ori | --> | 0 | 3371 | 589 | rep_origin | no |
| Ft# | Label or Note | Dir | Sta | End | Len | type | orf? |
|---|---|---|---|---|---|---|---|
| 0 | nd | --> | 0 | 2688 | 2688 | source | no |
| 1 | L:pBR322ori-F | --> | 117 | 137 | 20 | primer_bind | no |
| 2 | L:L4440 | --> | 370 | 388 | 18 | primer_bind | no |
| 3 | L:CAP binding si | --> | 504 | 526 | 22 | protein_bind | no |
| 4 | L:lac promoter | --> | 540 | 571 | 31 | promoter | no |
| 5 | L:lac operator | --> | 578 | 595 | 17 | protein_bind | no |
| 6 | L:M13/pUC Revers | --> | 583 | 606 | 23 | primer_bind | no |
| 7 | L:M13 rev | --> | 602 | 619 | 17 | primer_bind | no |
| 8 | L:M13 Reverse | --> | 602 | 619 | 17 | primer_bind | no |
| 9 | L:BsaI-F | --> | 631 | 638 | 7 | misc_feature | no |
| 10 | L:oh-f3-f4-F | --> | 638 | 642 | 4 | misc_feature | no |
| 11 | L:L2U3H03 | --> | 642 | 679 | 37 | misc_feature | no |
| 12 | L:oh-f7-v-F | --> | 679 | 683 | 4 | misc_feature | no |
| 13 | L:BsaI-R | --> | 683 | 690 | 7 | misc_feature | no |
| 14 | L:M13 Forward | <-- | 690 | 708 | 18 | primer_bind | no |
| 15 | L:M13 fwd | <-- | 690 | 707 | 17 | primer_bind | no |
| 16 | L:M13/pUC Forwar | <-- | 699 | 722 | 23 | primer_bind | no |
| 17 | L:pRS-marker | <-- | 915 | 935 | 20 | primer_bind | no |
| 18 | L:pGEX 3' | --> | 1034 | 1057 | 23 | primer_bind | no |
| 19 | L:pBRforEco | <-- | 1094 | 1113 | 19 | primer_bind | no |
| 20 | L:AmpR promoter | --> | 1180 | 1285 | 105 | promoter | no |
| 21 | L:AmpR | --> | 1285 | 2146 | 861 | CDS | yes |
| 22 | L:Amp-R | <-- | 1503 | 1523 | 20 | primer_bind | no |
| 23 | L:ori | --> | 0 | 2688 | 589 | rep_origin | no |
## search by label and return the location
def get_positions_by_label(record, label) :
feature_list = record.features
pos = {'start': None, 'end': None}
for feature in feature_list:
if "label" in feature.qualifiers:
if feature.qualifiers['label'] == [label]:
pos['start'] = int(feature.location.start)
pos['end'] = int(feature.location.end)
return pos
def get_record_between_labels(record, label1, label2) :
pos = {'start': get_positions_by_label(record, label1)['start'], 'end': get_positions_by_label(record, label2)['end']}
if pos['start'] == None or pos['end'] == None:
print("label not found")
return None
else:
return record[pos['start']:pos['end']]
target_record = get_record_between_labels(egfp, "BsaI-F", "BsaI-R")
# print("pos:", target_record)
display(target_record.figure())
target_record.seqDseqrecord(-742)
GGTCTCAGTCAATGGTGAGCAAGGGCGAGGAGCTGTTCACCGGGGTGGTGCCCATCCTGGTCGAGCTGGACGGCGACGTAAACGGCCACAAGTTCAGCGTGTCCGGCGAGGGCGAGGGCGATGCCACCTACGGCAAGCTGACCCTGAAGTTCATCTGCACCACCGGCAAGCTGCCCGTGCCCTGGCCCACCCTCGTGACCACCCTGACCTACGGCGTGCAGTGCTTCAGCCGCTACCCCGACCACATGAAGCAGCACGACTTCTTCAAGTCCGCCATGCCCGAAGGCTACGTCCAGGAGCGCACCATCTTCTTCAAGGACGACGGCAACTACAAGACCCGCGCCGAGGTGAAGTTCGAGGGCGACACCCTGGTGAACCGCATCGAGCTGAAGGGCATCGACTTCAAGGAGGACGGCAACATCCTGGGGCACAAGCTGGAGTACAACTACAACAGCCACAACGTCTATATCATGGCCGACAAGCAGAAGAACGGCATCAAGGTGAACTTCAAGATCCGCCACAACATCGAGGACGGCAGCGTGCAGCTCGCCGACCACTACCAGCAGAACACCCCCATCGGCGACGGCCCCGTGCTGCTGCCCGACAACCACTACCTGAGCACCCAGTCCGCCCTGAGCAAAGACCCCAACGAGAAGCGCGATCACATGGTCCTGCTGGAGTTCGTGACCGCCGCCGGGATCACTCTCGGCATGGACGAGCTGTACAAGTAAGGGATGAGACC
CCAGAGTCAGTTACCACTCGTTCCCGCTCCTCGACAAGTGGCCCCACCACGGGTAGGACCAGCTCGACCTGCCGCTGCATTTGCCGGTGTTCAAGTCGCACAGGCCGCTCCCGCTCCCGCTACGGTGGATGCCGTTCGACTGGGACTTCAAGTAGACGTGGTGGCCGTTCGACGGGCACGGGACCGGGTGGGAGCACTGGTGGGACTGGATGCCGCACGTCACGAAGTCGGCGATGGGGCTGGTGTACTTCGTCGTGCTGAAGAAGTTCAGGCGGTACGGGCTTCCGATGCAGGTCCTCGCGTGGTAGAAGAAGTTCCTGCTGCCGTTGATGTTCTGGGCGCGGCTCCACTTCAAGCTCCCGCTGTGGGACCACTTGGCGTAGCTCGACTTCCCGTAGCTGAAGTTCCTCCTGCCGTTGTAGGACCCCGTGTTCGACCTCATGTTGATGTTGTCGGTGTTGCAGATATAGTACCGGCTGTTCGTCTTCTTGCCGTAGTTCCACTTGAAGTTCTAGGCGGTGTTGTAGCTCCTGCCGTCGCACGTCGAGCGGCTGGTGATGGTCGTCTTGTGGGGGTAGCCGCTGCCGGGGCACGACGACGGGCTGTTGGTGATGGACTCGTGGGTCAGGCGGGACTCGTTTCTGGGGTTGCTCTTCGCGCTAGTGTACCAGGACGACCTCAAGCACTGGCGGCGGCCCTAGTGAGAGCCGTACCTGCTCGACATGTTCATTCCCTACTCTGG
Dseq(-742)
GGTC..GACC
CCAG..CTGGfrom pydna.readers import read
from primers import create, primers
from pydna.all import pcr
label1 = "BsaI-F"
label2 = "BsaI-R"
promoter_target = get_record_between_labels(promoter, label1, label2)
rbs_target = get_record_between_labels(rbs, label1, label2)
egfp_target = get_record_between_labels(egfp, label1, label2)
terminator_target = get_record_between_labels(terminator, label1, label2)
fwd, rev = create(str(promoter_target.seq))
promoter_pcr_product = pcr(fwd.seq, rev.seq, promoter)
display(promoter_pcr_product.figure())
fwd, rev = create(str(rbs_target.seq))
rbs_pcr_product = pcr(fwd.seq, rev.seq, rbs)
display(rbs_pcr_product.figure())
fwd, rev = create(str(egfp_target.seq))
egfp_pcr_product = pcr(fwd.seq, rev.seq, egfp)
display(egfp_pcr_product.figure())
fwd, rev = create(str(terminator_target.seq))
terminator_pcr_product = pcr(fwd.seq, rev.seq, terminator)
display(terminator_pcr_product.figure())5GGTCTCAAAGCTTGACG...CTAGCCTCCAGAGACC3
||||||||||||||||
3GATCGGAGGTCTCTGG5
5GGTCTCAAAGCTTGACG3
|||||||||||||||||
3CCAGAGTTTCGAACTGC...GATCGGAGGTCTCTGG55GGTCTCACTCCAGCTG...GAGGAGAAATAGTCATGAGACC3
||||||||||||||||||||||
3CTCCTCTTTATCAGTACTCTGG5
5GGTCTCACTCCAGCTG3
||||||||||||||||
3CCAGAGTGAGGTCGAC...CTCCTCTTTATCAGTACTCTGG55GGTCTCAGTCAATGGTGA...TACAAGTAAGGGATGAGACC3
||||||||||||||||||||
3ATGTTCATTCCCTACTCTGG5
5GGTCTCAGTCAATGGTGA3
||||||||||||||||||
3CCAGAGTCAGTTACCACT...ATGTTCATTCCCTACTCTGG55GGTCTCAGGGATAGCG...TTGTTGAGCGAATGAGACC3
|||||||||||||||||||
3AACAACTCGCTTACTCTGG5
5GGTCTCAGGGATAGCG3
||||||||||||||||
3CCAGAGTCCCTATCGC...AACAACTCGCTTACTCTGG5from Bio.Restriction import BsaI
from pydna.assembly import Assembly
from pydna.common_sub_strings import terminal_overlap
promoter_cut_product = promoter_pcr_product.cut(BsaI)
print(len(promoter_cut_product))
promoter_cut_product[1].name = "promoter"
display(promoter_cut_product[1].figure())
rbs_cut_product = rbs_pcr_product.cut(BsaI)
print(len(rbs_cut_product))
rbs_cut_product[1].name = "rbs"
display(rbs_cut_product[1].figure())
egfp_cut_product = egfp_pcr_product.cut(BsaI)
print(len(egfp_cut_product))
egfp_cut_product[1].name = "egfp"
display(egfp_cut_product[1].figure())
terminator_cut_product = terminator_pcr_product.cut(BsaI)
print(len(terminator_cut_product))
terminator_cut_product[1].name = "terminator"
display(terminator_cut_product[1].figure())
asm = Assembly((promoter_cut_product[1], rbs_cut_product[1], egfp_cut_product[1], terminator_cut_product[1]), algorithm = terminal_overlap, limit = 4)
candidate = asm.assemble_linear()
print(len(candidate))
candidate[0].figure()
3
3
3
3
1Dseqrecord(-43)
AAGCTTGACGGCTAGCTCAGTCCTAGGTACAGTGCTAGC
AACTGCCGATCGAGTCAGGATCCATGTCACGATCGGAGG
Dseqrecord(-107)
CTCCAGCTGTCACCGGATGTGCTTTCCGGTCTGATGAGTCCGTGAGGACGAAACAGCCTCTACAAATAATTTTGTTTAATCTAGAGATTAAAGAGGAGAAATA
TCGACAGTGGCCTACACGAAAGGCCAGACTACTCAGGCACTCCTGCTTTGTCGGAGATGTTTATTAAAACAAATTAGATCTCTAATTTCTCCTCTTTATCAGT
Dseqrecord(-728)
GTCAATGGTGAGCAAGGGCGAGGAGCTGTTCACCGGGGTGGTGCCCATCCTGGTCGAGCTGGACGGCGACGTAAACGGCCACAAGTTCAGCGTGTCCGGCGAGGGCGAGGGCGATGCCACCTACGGCAAGCTGACCCTGAAGTTCATCTGCACCACCGGCAAGCTGCCCGTGCCCTGGCCCACCCTCGTGACCACCCTGACCTACGGCGTGCAGTGCTTCAGCCGCTACCCCGACCACATGAAGCAGCACGACTTCTTCAAGTCCGCCATGCCCGAAGGCTACGTCCAGGAGCGCACCATCTTCTTCAAGGACGACGGCAACTACAAGACCCGCGCCGAGGTGAAGTTCGAGGGCGACACCCTGGTGAACCGCATCGAGCTGAAGGGCATCGACTTCAAGGAGGACGGCAACATCCTGGGGCACAAGCTGGAGTACAACTACAACAGCCACAACGTCTATATCATGGCCGACAAGCAGAAGAACGGCATCAAGGTGAACTTCAAGATCCGCCACAACATCGAGGACGGCAGCGTGCAGCTCGCCGACCACTACCAGCAGAACACCCCCATCGGCGACGGCCCCGTGCTGCTGCCCGACAACCACTACCTGAGCACCCAGTCCGCCCTGAGCAAAGACCCCAACGAGAAGCGCGATCACATGGTCCTGCTGGAGTTCGTGACCGCCGCCGGGATCACTCTCGGCATGGACGAGCTGTACAAGTAA
TACCACTCGTTCCCGCTCCTCGACAAGTGGCCCCACCACGGGTAGGACCAGCTCGACCTGCCGCTGCATTTGCCGGTGTTCAAGTCGCACAGGCCGCTCCCGCTCCCGCTACGGTGGATGCCGTTCGACTGGGACTTCAAGTAGACGTGGTGGCCGTTCGACGGGCACGGGACCGGGTGGGAGCACTGGTGGGACTGGATGCCGCACGTCACGAAGTCGGCGATGGGGCTGGTGTACTTCGTCGTGCTGAAGAAGTTCAGGCGGTACGGGCTTCCGATGCAGGTCCTCGCGTGGTAGAAGAAGTTCCTGCTGCCGTTGATGTTCTGGGCGCGGCTCCACTTCAAGCTCCCGCTGTGGGACCACTTGGCGTAGCTCGACTTCCCGTAGCTGAAGTTCCTCCTGCCGTTGTAGGACCCCGTGTTCGACCTCATGTTGATGTTGTCGGTGTTGCAGATATAGTACCGGCTGTTCGTCTTCTTGCCGTAGTTCCACTTGAAGTTCTAGGCGGTGTTGTAGCTCCTGCCGTCGCACGTCGAGCGGCTGGTGATGGTCGTCTTGTGGGGGTAGCCGCTGCCGGGGCACGACGACGGGCTGTTGGTGATGGACTCGTGGGTCAGGCGGGACTCGTTTCTGGGGTTGCTCTTCGCGCTAGTGTACCAGGACGACCTCAAGCACTGGCGGCGGCCCTAGTGAGAGCCGTACCTGCTCGACATGTTCATTCCCT
Dseqrecord(-45)
GGGATAGCGTGACCGGCGCATCGGTCACGCTATTTGTTGAG
ATCGCACTGGCCGCGTAGCCAGTGCGATAAACAACTCGCTT
promoter| 4
\/
/\
4|rbs| 4
\/
/\
4|egfp| 4
\/
/\
4|terminatorasm = Assembly((promoter_cut_product[0], rbs_cut_product[1], egfp_cut_product[1], terminator_cut_product[1]), algorithm = terminal_overlap, limit = 4)
candidate = asm.assemble_linear()
print(len(candidate))0